Posts
An article from the Nature.
I used {ggh4x} package for the first time, to modify y-axis ticks.
Selected article:
Title: The KEYNOTE-811 trial of dual PD-1 and HER2 blockade in HER2-positive gastric cancer
Journal: Nature
Authors: Janjigian YY, Kawazoe A, Yañez P et al.
Year: 2021
PMID: 34912120
DOI: 10.1038/s41586-021-04161-3
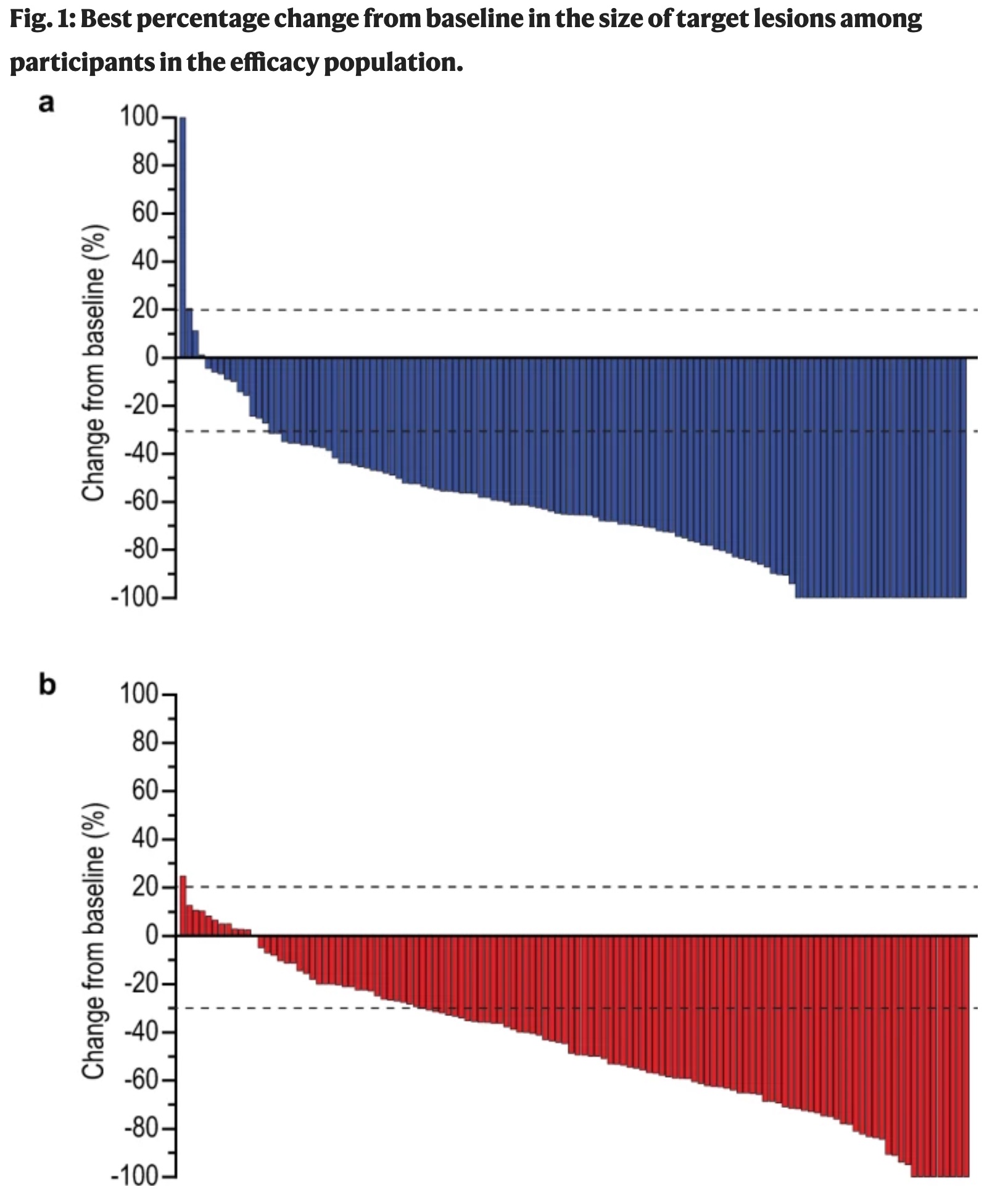
The original figure
Import libraries
library(tidyverse)
library(scales)
library(fabricatr) # to fabricate fake data
library(patchwork) # to combine plots
library(ggh4x) # to modify y-axis ticks
theme_set(theme_light(base_family = "Helvetica Neue"))
Prepare fabricated data
set.seed(2022)
# Pembrolizumab group
pembro_progressive <- fabricate(
N = 1,
main_group = "Pembro",
subgroup = "progressive",
pre_treatment = 100,
post_treatment = 240)
pembro_stabil <- fabricate(
N = 15,
main_group = "Pembro",
subgroup = "stabil",
pre_treatment = 100,
post_treatment = round(runif(N, 70, 120))
)
pembro_partial <- fabricate(
N = 84,
main_group = "Pembro",
subgroup = "partial",
pre_treatment = 100,
post_treatment = round(runif(N, 1, 70))
)
pembro_complete <- fabricate(
N = 28,
main_group = "Pembro",
subgroup = "complete",
pre_treatment = 100,
post_treatment = 0)
set.seed(2022)
data_pembro <- bind_rows(pembro_progressive, pembro_stabil, pembro_partial, pembro_complete) %>%
mutate(response_rate = pmin(100, 100 * (post_treatment - pre_treatment)/pre_treatment)) %>%
filter(response_rate != 0) %>%
sample_n(124)
# Placebo group
set.seed(2022)
placebo_progressive <- fabricate(
N = 1,
main_group = "Placebo",
subgroup = "progressive",
pre_treatment = 100,
post_treatment = 128)
placebo_stabil <- fabricate(
N = 38,
main_group = "Placebo",
subgroup = "stabil",
pre_treatment = 100,
post_treatment = round(runif(N, 70, 120))
)
placebo_partial <- fabricate(
N = 77,
main_group = "Placebo",
subgroup = "partial",
pre_treatment = 100,
post_treatment = round(runif(N, 1, 70))
)
placebo_complete <- fabricate(
N = 9,
main_group = "Placebo",
subgroup = "complete",
pre_treatment = 100,
post_treatment = 0)
set.seed(2022)
data_placebo <- bind_rows(placebo_progressive, placebo_stabil, placebo_partial, placebo_complete) %>%
mutate(response_rate = 100 * (post_treatment - pre_treatment)/pre_treatment) %>%
sample_n(122)
# combine datasets
combined_dataset <- bind_rows(data_pembro, data_placebo) %>%
as_tibble () %>%
select(-ID) %>%
mutate(patient_ID = row_number(), .before = "main_group")
A part of fake dataset
set.seed(2022)
combined_dataset %>%
sample_n(10)
## # A tibble: 10 × 6
## patient_ID main_group subgroup pre_treatment post_treatment response_rate
## <int> <chr> <chr> <dbl> <dbl> <dbl>
## 1 228 Placebo stabil 100 78 -22
## 2 179 Placebo partial 100 57 -43
## 3 206 Placebo partial 100 21 -79
## 4 55 Pembro partial 100 19 -81
## 5 75 Pembro partial 100 66 -34
## 6 196 Placebo partial 100 58 -42
## 7 6 Pembro partial 100 33 -67
## 8 191 Placebo partial 100 28 -72
## 9 244 Placebo partial 100 59 -41
## 10 220 Placebo partial 100 11 -89
Possible strategy:
Facet approach is OK but I will prepare two separate plots.
I decide this (faceted or separate plots) based on the complexity of axis etc.
However, We can use the @drob rule to decide.
R codes for the figure
# Pembro plot
plot_pembro <- combined_dataset %>%
filter(main_group == "Pembro") %>%
mutate(patient_ID = reorder(patient_ID, -response_rate)) %>%
ggplot(aes(patient_ID, response_rate)) +
geom_col(fill = "#2A4093", position = "dodge", color = "black") +
scale_y_continuous(breaks = seq(-100, 100, 20),
minor_breaks = seq(-100, 100, by = 10),
guide = "axis_minor",
limits = c(-100, 100),
expand = c(0, 0)) +
scale_x_discrete(expand = expansion(add = c(1,2))) +
geom_hline(yintercept = c(-30, 20), lty = 2, size = .70) +
geom_hline(yintercept = 0, lty = 1, size = .80) +
labs(x = "",
y = "Change from baseline (%)") +
theme(axis.text.x = element_blank(),
axis.text.y = element_text(size = 14),
axis.title.y = element_text(size = 14),
panel.grid = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_line(color = "black", size = 1),
axis.ticks.length.y = unit(.3, "cm"),
panel.border = element_blank(),
axis.line.y = element_line(color = "black"),
ggh4x.axis.ticks.length.minor = rel(0.5))
# Placebo plot (only filter and color hex was changed)
plot_placebo <- combined_dataset %>%
filter(main_group == "Placebo") %>%
mutate(patient_ID = reorder(patient_ID, -response_rate)) %>%
ggplot(aes(patient_ID, response_rate)) +
geom_col(fill = "#DF001B", position = "dodge", color = "black") +
scale_y_continuous(breaks = seq(-100, 100, 20),
minor_breaks = seq(-100, 100, by = 10),
guide = "axis_minor",
limits = c(-100, 100),
expand = c(0, 0)) +
scale_x_discrete(expand = expansion(add = c(1,2))) +
geom_hline(yintercept = c(-30, 20), lty = 2, size = .70) +
geom_hline(yintercept = 0, lty = 1, size = .80) +
labs(x = "",
y = "Change from baseline (%)") +
theme(axis.text.x = element_blank(),
axis.text.y = element_text(size = 14),
axis.title.y = element_text(size = 14),
panel.grid = element_blank(),
axis.ticks.x = element_blank(),
axis.ticks.y = element_line(color = "black", size = 1),
axis.ticks.length.y = unit(.3, "cm"),
panel.border = element_blank(),
axis.line.y = element_line(color = "black"),
ggh4x.axis.ticks.length.minor = rel(0.5))
# Combine plots
final_replica <- plot_pembro / plot_placebo +
plot_annotation(title = "Fig. 1: Best percentage change from baseline in the size of target lesions among \nparticipants in the efficacy population.",
tag_levels = "a") &
theme(plot.tag = element_text(size = 16, face = "bold"),
plot.title = element_text(face = "bold", family = "Palatino LT Black", hjust = 0, lineheight = 1.5))
## SAVE FIGURE
ggsave(final_replica,
file =file.path ("w8_replica.jpg"),
dpi = 300,
width = 7.4,
height = 9.2)
Final replica
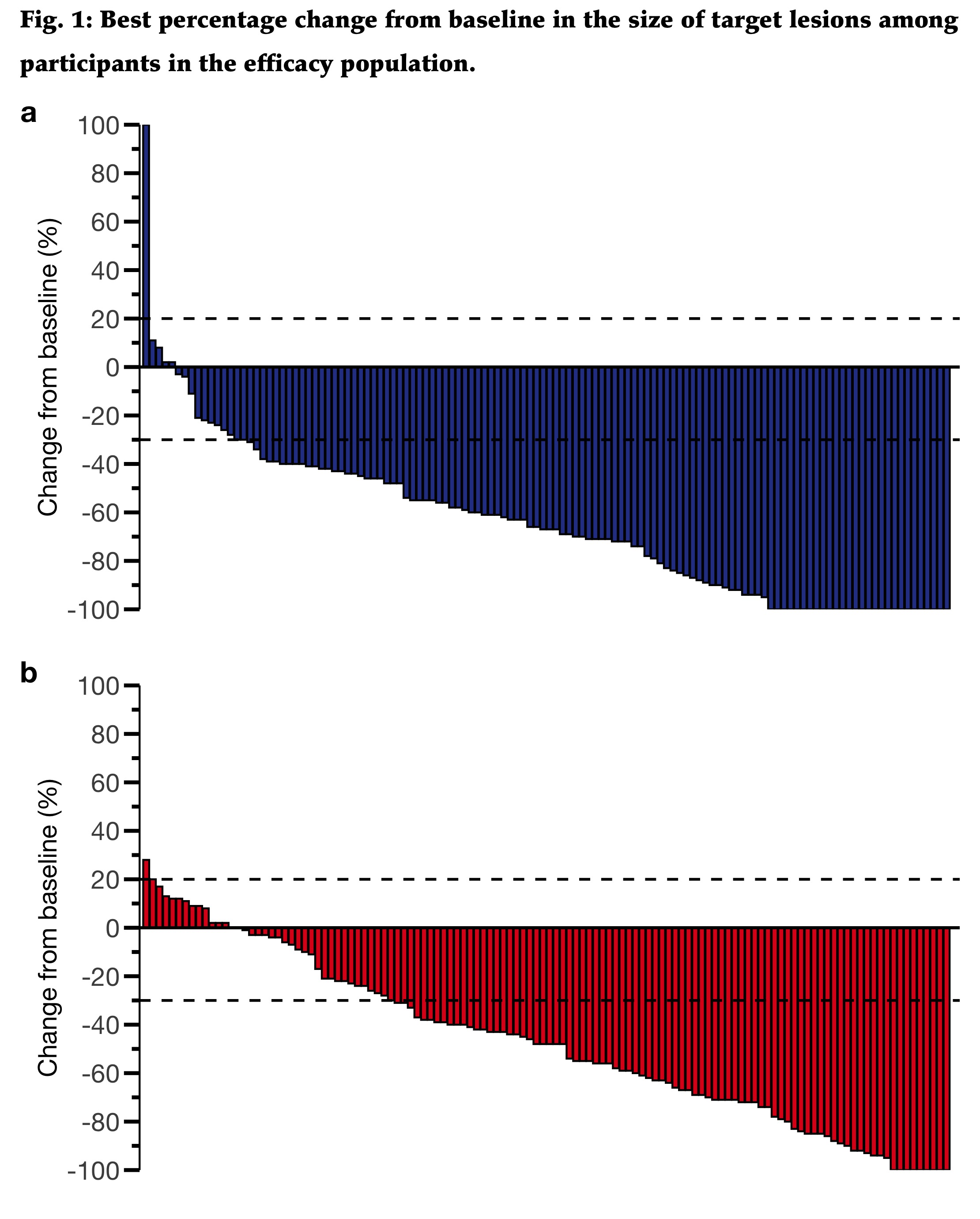
Some personal comments:
I m not sure whether this is an informative figure. I would prefer (based on domain knowledge) using bar graph to show the frequencies of the response group.
This one does not allow to compare the groups.
Citation
Ali Guner (Apr 14, 2022) Week-8. Retrieved from https://datavizmed.com/blog/2022-04-14-week-8/
@misc{ 2022-week-8,
author = { Ali Guner },
title = { Week-8 },
url = { https://datavizmed.com/blog/2022-04-14-week-8/ },
year = { 2022 }
updated = { Apr 14, 2022 }
}