Posts
a question from twitter by JulienMouchnino,
can we prepare grouped summary table with flextable?
Although this is not related to medicine, can be used in any table, therefore I shared it here.
The original table
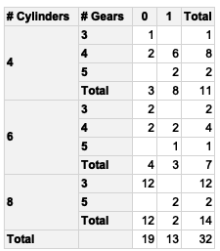
Import libraries
library(tidyverse)
library(flextable)
library(officer)
Possible strategy:
Although gt() packages provide row-based, column-based and all-based summary table, I prefer to complete summary as I desire.
Then, I use flextable() to modify my summarized tibble.
Summary table
summary_col <- mtcars %>%
summarise(nb = n(), .by = c(cyl, gear, am)) %>%
arrange(cyl, gear, am) %>%
pivot_wider(names_from = "am",
values_from = "nb",
names_prefix = "am_",
values_fill = 0) %>%
mutate(total = am_0 + am_1)
summary_row <- summary_col %>%
summarise(across(am_0:total, sum), .by = c("cyl")) %>%
add_column(gear = "Total", .after = "cyl")
summary_all <- summary_row %>%
summarise(across(am_0:total, sum)) %>%
add_column(gear = "", .before = 1) %>%
add_column(cyl = "Total", .before = 1)
summary_table <- bind_rows(
summary_row %>%
mutate(across(everything(), as.character)),
summary_col %>%
mutate(across(everything(), as.character)),
summary_all %>%
mutate(across(everything(), as.character))) %>%
arrange(cyl, gear) %>%
mutate(across(am_0:total, ~str_replace_all(., "0", NA_character_)))
Convert summary table to flextable
col_table_border <- "#a6a6a6"
col_table_bg <- "#e6e6e6"
flex_summary <- summary_table %>%
flextable() %>%
border_remove() %>%
set_table_properties(width = 1, layout = "autofit") %>%
hline(part = "all", border = fp_border(color = col_table_border)) %>%
vline(part = "all", border = fp_border(color = col_table_border)) %>%
border_outer(border = fp_border(color = col_table_border), part = "all") %>%
set_header_labels(cyl = "# Cylinders",
gear = "# Gears",
am_0 = "0",
am_1 = "1",
total = "Total") %>%
fix_border_issues() %>%
merge_v(j = "cyl") %>%
bold(part = "header") %>%
bold(part = "body", j = 1:2) %>%
align(part = "header", align = "left") %>%
align(j = 3:5, align = "right") %>%
bg(part = "header", bg = col_table_bg) %>%
bg(part = "body", j = 1:2, bg = col_table_bg) %>%
font(fontname = "Helvetica")
Save flextable object as a word file
save_as_docx(flex_summary,
path = here::here("content", "blog", "2023-10-13-week-11", paste0("flex_summary", ".docx")))
Final replica
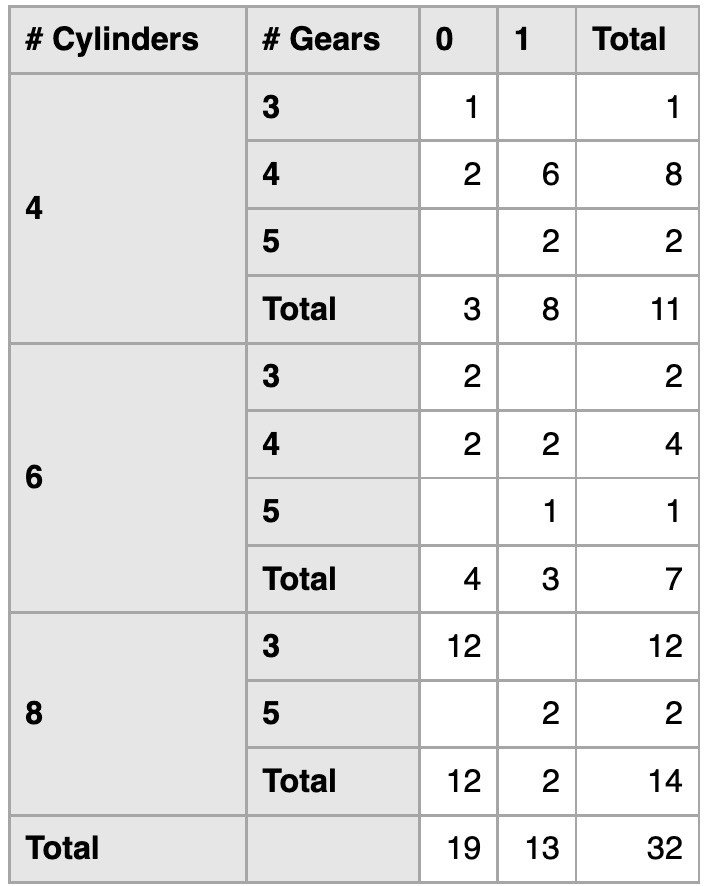
Citation
For attribution, please cite this work as
Ali Guner (Oct 13, 2023) W11: Custom summary table. Retrieved from https://datavizmed.com/blog/2023-10-13-week-11/
BibTeX citation
@misc{ 2023-w11-custom-summary-table,
author = { Ali Guner },
title = { W11: Custom summary table },
url = { https://datavizmed.com/blog/2023-10-13-week-11/ },
year = { 2023 }
updated = { Oct 13, 2023 }
}