Posts
This week’s article is from the University of Pennsylvania.
Selected article:
Title: Disparities in Presentation, Treatment, and Survival in Anaplastic Thyroid Cancer
Journal: Annals of Surgical Oncology
Authors: Ginzberg SP, Gasior JA, Passman JE et al.
Year: 2023
PMID: 37474696
DOI: 10.1245/s10434-023-13945-y
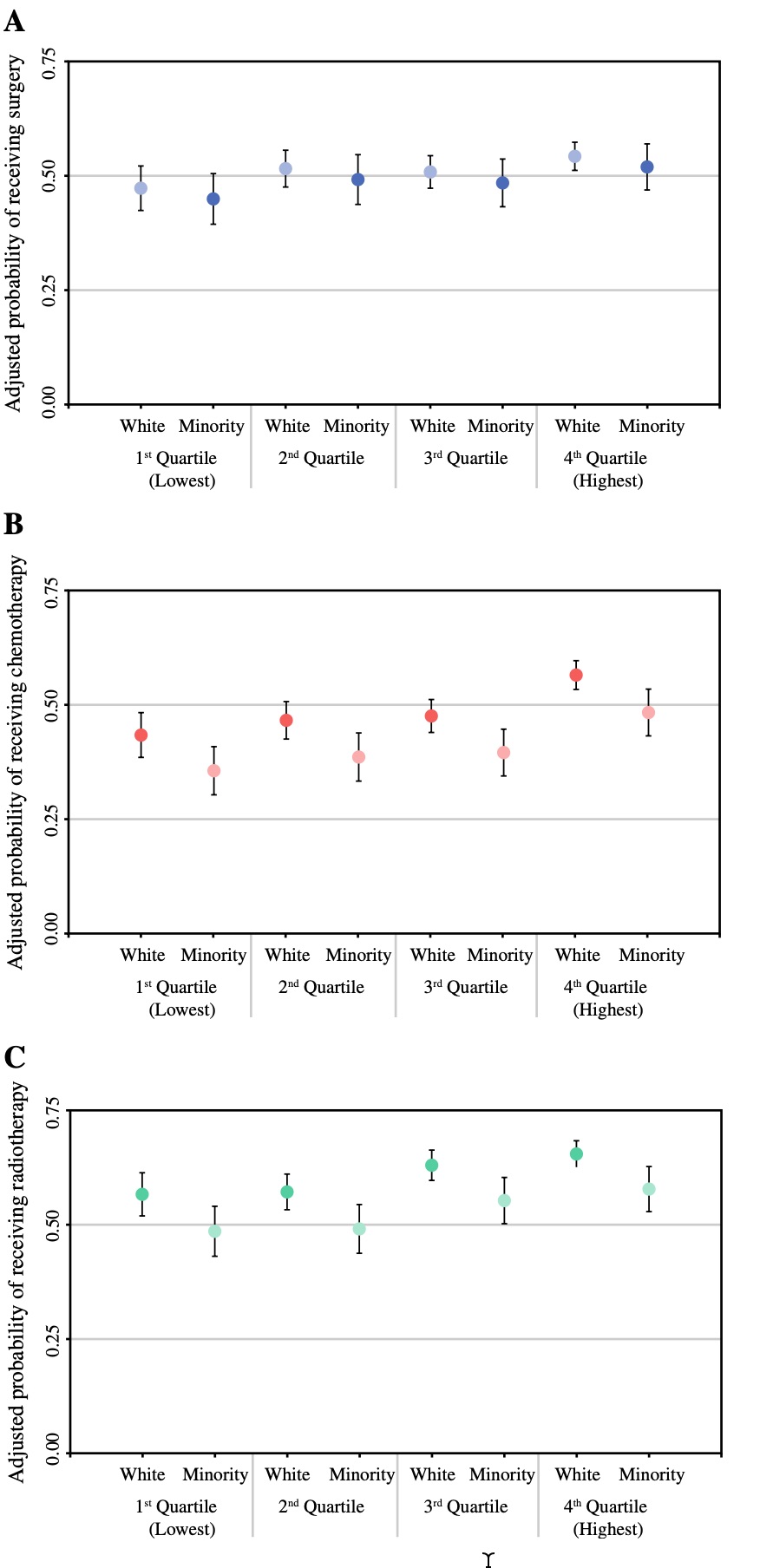
The original figure
Import libraries
# Define the required package names
packages <- c("tidyverse", "scales", "fabricatr", "glue",
"ggtext", "cowplot")
# Function to check, install, and load packages
load_packages <- function(packages) {
for (package in packages) {
if (!require(package, character.only = TRUE)) {
install.packages(package)
library(package, character.only = TRUE)
} else {
# suppressPackageStartupMessages(library(package, character.only = TRUE))
library(package, character.only = TRUE)
}
}
}
load_packages(packages)
theme_set(theme_light(base_family = "Times New Roman"))
Prepare fabricated data
generate_thyroid_data <- function(N, main_group, subgroup, prob_surgery, prob_chemo, prob_radio) {
set.seed(2023)
fabricate(
N = N,
main_group = main_group,
subgroup = subgroup,
surgery = draw_binary(N = N, prob = prob_surgery),
chemotherapy = draw_binary(N = N, prob = prob_chemo),
radiotherapy = draw_binary(N = N, prob = prob_radio)
)
}
## Non-hispanic white
thyroid_white_q1 <- generate_thyroid_data(N = 498, main_group = "non_hispanic", subgroup = "Q1", 0.47, 0.45, 0.54)
thyroid_white_q2 <- generate_thyroid_data(N = 834, main_group = "non_hispanic", subgroup = "Q2", 0.52, 0.47, 0.55)
thyroid_white_q3 <- generate_thyroid_data(N = 1037, main_group = "non_hispanic", subgroup = "Q3", 0.51, 0.47, 0.61)
thyroid_white_q4 <- generate_thyroid_data(N = 1430, main_group = "non_hispanic", subgroup = "Q4", 0.54, 0.56, 0.63)
## Minority
thyroid_minority_q1 <- generate_thyroid_data(N = 263, main_group = "minority", subgroup = "Q1", 0.45, 0.32, 0.49)
thyroid_minority_q2 <- generate_thyroid_data(N = 204, main_group = "minority", subgroup = "Q2", 0.49, 0.34, 0.48)
thyroid_minority_q3 <- generate_thyroid_data(N = 217, main_group = "minority", subgroup = "Q3", 0.48, 0.35, 0.54)
thyroid_minority_q4 <- generate_thyroid_data(N = 277, main_group = "minority", subgroup = "Q4", 0.52, 0.48, 0.55)
## Combine 8 datasets
combined_dataset <- bind_rows(
thyroid_white_q1,
thyroid_white_q2,
thyroid_white_q3,
thyroid_white_q4,
thyroid_minority_q1,
thyroid_minority_q2,
thyroid_minority_q3,
thyroid_minority_q4) %>%
as_tibble() %>%
mutate(across(surgery:radiotherapy, ~if_else(. == 1, "yes", "no")))
A part of fake dataset
set.seed(2023)
combined_dataset %>%
sample_n(10)
## # A tibble: 10 × 6
## ID main_group subgroup surgery chemotherapy radiotherapy
## <chr> <chr> <chr> <chr> <chr> <chr>
## 1 0110 non_hispanic Q4 yes yes yes
## 2 0053 non_hispanic Q3 yes yes no
## 3 0660 non_hispanic Q3 no yes no
## 4 0985 non_hispanic Q4 yes no yes
## 5 368 non_hispanic Q2 yes no yes
## 6 0233 non_hispanic Q3 yes no yes
## 7 1264 non_hispanic Q4 yes yes yes
## 8 659 non_hispanic Q2 yes yes yes
## 9 461 non_hispanic Q2 yes no no
## 10 095 minority Q4 no no no
Possible strategy:
I ll go with faceted figure.
I ll write a custom function for one facet (quartiles), and I ll use the function for all three treatment types.
The most difficult (and useless) part was to add gray vertical lines between facet names. I used cowplot::draw_line() to add manuel lines. This solution solved the problem but seems problematic because the coordinates should be defined manually and checked individually.
Calculate percentages and CIs
cis_dataset <- combined_dataset %>%
pivot_longer(cols = c(surgery:radiotherapy)) %>%
group_by(main_group, subgroup, name) %>%
summarise(count_yes = sum(value == "yes"),
n = n(),
prop_yes = count_yes / n,
conf_interval = list(prop.test(count_yes, n)$conf.int),
.groups = "drop") %>%
mutate(lower_bound = map_dbl(conf_interval, ~ .x[1]),
upper_bound = map_dbl(conf_interval, ~ .x[2])) %>%
select(-conf_interval)
R codes for the figure
quartile_names <- as_labeller(c( # for facet names
"Q1" = "1^st^ Quartile<br>(Lowest)",
"Q2" = "2^nd^ Quartile",
"Q3" = "3^rd^ Quartile",
"Q4" = "4^th^ Quartile<br>(Highest)"
))
draw_myplot <- function(my_name){
text_y <- glue::glue("Adjusted probability of receiving {my_name}") # to add y-axis title dynamically
selected_colors <- case_when( # to define colors dynamically
my_name == "surgery" ~ c("#A6B5D8", "#5069B8"),
my_name == "chemotherapy" ~ c("#E3665E", "#F0B1B2"),
my_name == "radiotherapy" ~ c("#75CEA5", "#B5E7D2"))
initial_plot <- cis_dataset %>%
mutate(main_group = fct_relevel(main_group, "non_hispanic"),
name = fct_relevel(name, "surgery", "chemotherapy", "radiotherapy")) %>%
filter(name == {{my_name}}) %>%
ggplot(aes(main_group, prop_yes, color = main_group)) +
geom_errorbar(aes(ymin = lower_bound,
ymax = upper_bound), color = "black", width = 0.1) +
geom_point(size = 2.5) +
facet_wrap( ~ subgroup, ncol = 4, strip.position = "bottom", scales = "free_x", labeller = quartile_names) +
scale_y_continuous(limits = c(0, .75), breaks = seq(0, .75, .25), expand = c(0,0), sec.axis = dup_axis(breaks = NULL)) +
scale_x_discrete(labels = c("White", "Minority")) +
scale_color_manual(values = selected_colors) +
geom_hline(yintercept = 0.75, color = "black", linewidth = 1) +
labs(x = element_blank(),
y = text_y) +
theme(legend.position = "none",
text = element_text(color = "black"),
axis.text.y = element_text(angle = 90, hjust = 0.5),
axis.title.y.right = element_blank(),
axis.title.y = element_text(size = 10, margin = margin(r = 6)),
axis.line = element_line(color = "black"),
axis.ticks = element_line(color = "black", size = .5),
strip.placement = "outside",
strip.text = ggtext::element_markdown(color = "black", lineheight = 1),
strip.background = element_blank(),
panel.border = element_blank(),
panel.grid.minor.y = element_blank(),
panel.grid.major.x = element_blank(),
panel.spacing = unit(0, "lines"),
plot.margin = margin(1, 1, 0, .2, "cm"))
final_plot <- ggdraw() +
draw_plot(initial_plot) +
draw_line(x = c(0.29, 0.29), # to add vertical line between facet names manually
y = c(0.19, 0),
lwd = 0.5,
colour = "gray") +
draw_line(x = c(0.50, 0.50), # to add vertical line between facet names manually
y = c(0.19, 0),
lwd = 0.5,
colour = "gray") +
draw_line(x = c(0.71, 0.71), # to add vertical line between facet names manually
y = c(0.19, 0),
lwd = 0.5,
colour = "gray")
return(final_plot)
}
final_replica <- plot_grid( # to combine 3 plots and to add Tags
draw_myplot("surgery"),
draw_myplot("chemotherapy"),
draw_myplot("radiotherapy"),
labels = "AUTO",
label_fontface = "bold",
vjust = 1,
hjust = -.5,
ncol = 1)
### SAVE FIGURE
ggsave(final_replica,
file =file.path ("w12_replica.jpg"),
dpi = 300,
width = 5,
height = 10)
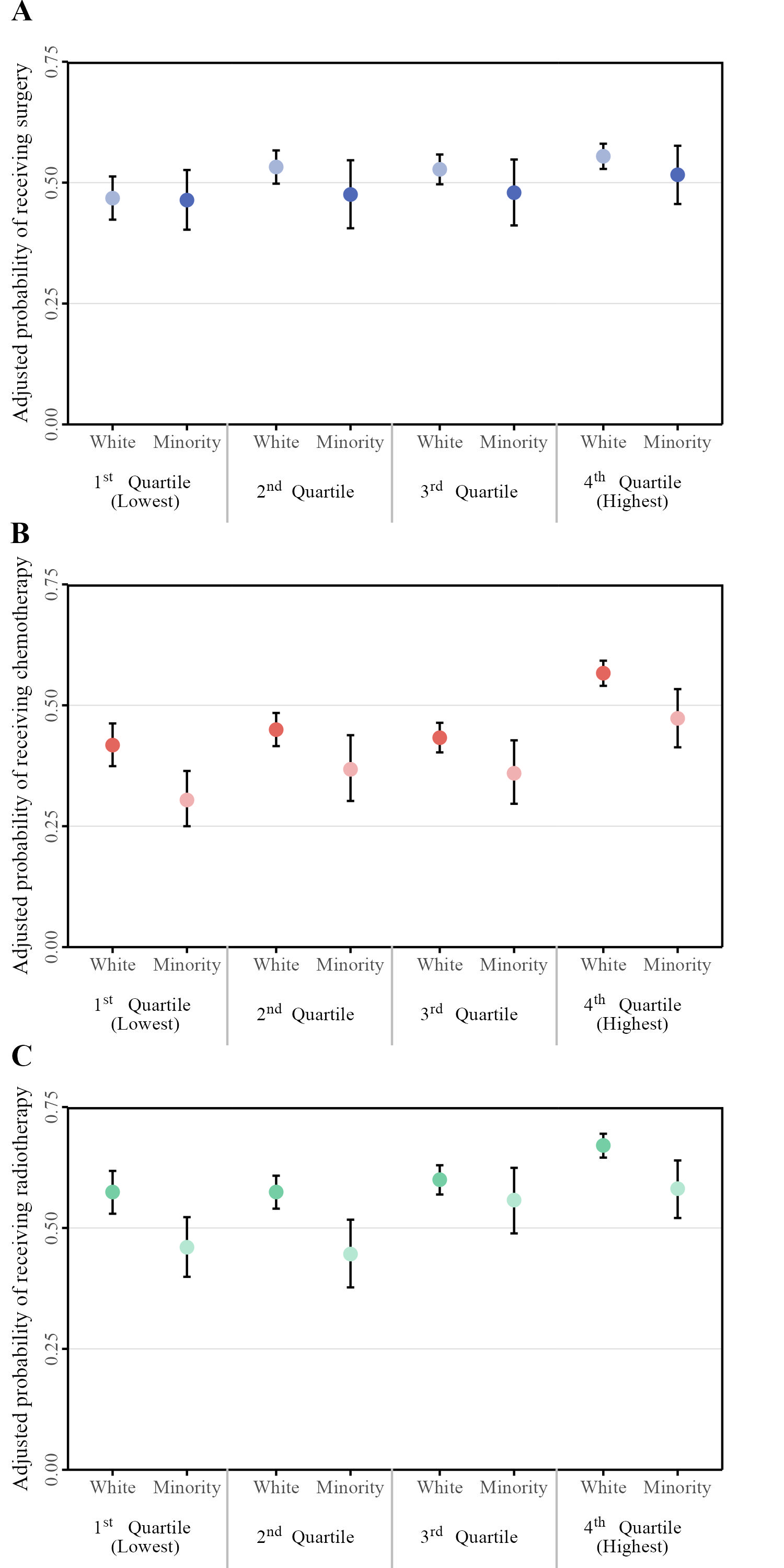
Final replica
Some personal comments:
Citation
Ali Guner (Oct 25, 2023) W12 - Errorbar plots with facets. Retrieved from https://datavizmed.com/blog/2023-10-25-week-12/
@misc{ 2023-w12-errorbar-plots-with-facets,
author = { Ali Guner },
title = { W12 - Errorbar plots with facets },
url = { https://datavizmed.com/blog/2023-10-25-week-12/ },
year = { 2023 }
updated = { Oct 25, 2023 }
}